UCLA researchers develop new, noninvasive AI method to inspect live cells and gain critical data
Researchers at the UCLA Samueli School of Engineering have discovered a new artificial intelligence-based method to discern the properties of live biological cells without destroying them. The advance could enable laboratories to conduct drug-safety screening faster and more efficiently while improving quality control for cell therapies.
The research was published today in Nature’s Scientific Reports.
"We want to know if a batch of live biological cells can be both viable and able to perform the functions we want them to. This noninvasive, AI-backed technique can infer the quality of those cells while keeping the entire batch intact," said study leader Neil Lin, an assistant professor of mechanical and aerospace engineering and member of the UCLA Broad Stem Cell Research Center. "We envision this method could be widely adopted by many academic and industrial cell biology labs. And it could be especially important in cell therapies, where the cells themselves are valuable."
Currently, cells are often characterized using antibody staining. This method requires cells to be isolated and dyed with fluorescent tags that light up when a target protein is present. Not only is such a process time-consuming — taking up to a full day to prepare, process and analyze the cells — it also kills the cells used for the analysis.
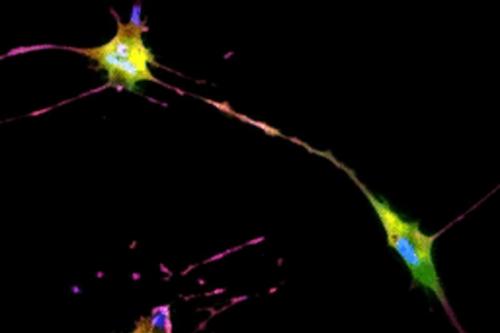
The UCLA researchers' new technique allows for instant cell assessments while keeping the entire batch of cells intact. Employing an AI-powered deep-learning model, cells are viewed and a snapshot taken under a light-based microscope, also known as a bright-field microscope. While the model is basically the same as one that has been used in the movie industry to alter and enhance images, such as artificially aging a movie character, the UCLA team adapted the model and trained it to infer and identify antibody-labeled fluorescent images of cells. The optimized model analyzes the subtle differences in size and shape of cells, qualities not readily visible to human eyes, and utilizes that information for predicting the levels of proteins present.
The processed image unveils information on existence of proteins and their whereabouts, much like a traditional stained sample would without sacrificing the cells. Moreover, this method may provide a more accurate assessment of the cells. While traditional staining methods can usually label a few different proteins, the new AI tool can predict as many proteins as the machine-learning model has been trained to identify.
The group focused on mesenchymal stem cells that are essential for biological tissue regeneration in cell therapies as they can orchestrate multiple types of cells to form new tissues. These cells also hold promise for treating various inflammatory disorders as they can modulate the human immune system. The researchers noted the imaging technique could be further refined if they can obtain more training data on a wide range of cell types and features, such as the age of cells and their changes following drug use.
The lead authors on the study are Sara Imboden, a visiting graduate student in mechanical engineering, and UCLA computer science graduate student Xuanqing Li. The other UCLA authors are bioengineering undergraduate student Brandon Lee, mechanical engineering graduate Marie Payne and Cho-Jui Hsieh — an assistant professor of computer science who works on machine-learning algorithms.
Lin leads the Living Soft Materials Research Laboratory at UCLA. He also has faculty appointment in the bioengineering department and is a member of the Institute for Quantitative and Computational Biosciences.
The research was supported by a UCLA SPORE in Prostate Cancer grant, the National Science Foundation and a UCLA Broad Stem Cell Research Center and UCLA California NanoSystems Institute Planning Award.
